Home
Unregistered user, login or register
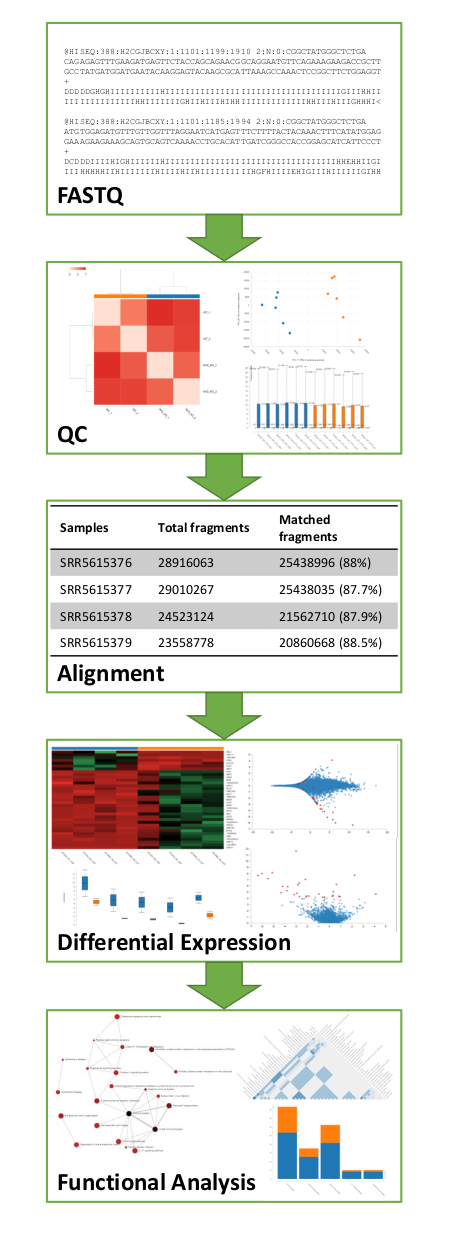
RaNA-Seq is an open bioinformatics tool for the quick analysis of RNA-Seq data. It performs a full analysis in minutes quantifying FASTQ files, calculating quality control metrics, running differential expression analyses and enabling the interpretation of results with functional analyses. Our analysis pipeline integrates cutting edge bioinformatics tools and simplify its application with a friendly Web interface designed for non-experienced users in these analyses. Each analysis can be customized setting up input parameters and applies generally accepted and reproducible protocols. Analysis results are presented as interactive graphics and reports, ready for their interpretation and publication.
Main features of RaNA-Seq:
- Full RNA-Seq analysis from the FASTQ file to the functional analysis of results
- Stand alone website without any software installation required
- Connected with the ENA repository
- Auto-configuration of input parameters oriented to non-experienced users
- Quality control of input samples with several graphs and metrics
- Customization of analyses with input parameters and several differential expression methods
- Functional over-representation analysis of results
- Gene Set Enrichment Analysis of results (GSEA)
- Generation of reports with full information about the analysis, ready for its inclusion in publications
- Presentation of results with interactive graphics (some examples of Differential expression results, functional enrichment results, Quality Control metrics)
- Generally accepted and reproducible protocols
Reference
Carlos Prieto, David Barrios, RaNA-Seq: interactive RNA-Seq analysis from FASTQ files to functional analysis, Bioinformatics, Volume 36, Issue 6, 15 March 2020, Pages 1955–1956, https://doi.org/10.1093/bioinformatics/btz854
