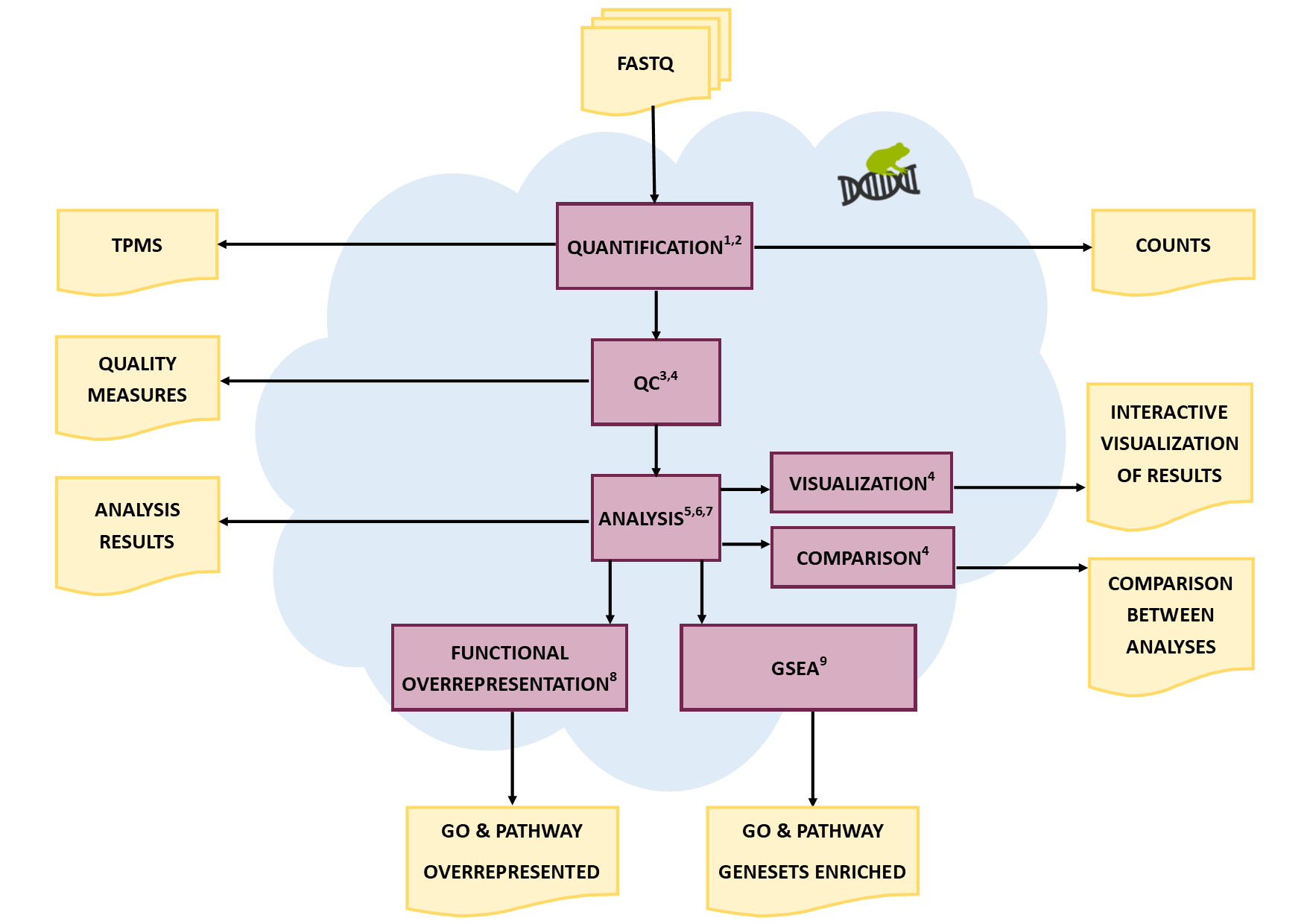
Pipeline
Unregistered user, login or register
- Chen,S. et al. (2018) Fastp: An ultra-fast all-in-one FASTQ preprocessor. In, Bioinformatics., pp. i884–i890.
- Patro,R. et al. (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods, 14, 417–419.
- Schulze,S.K. et al. (2012) SERE: Single-parameter quality control and sample comparison for RNA-Seq. BMC Genomics, 13.
- Barrios,D. and Prieto,C. (2018) RJSplot: Interactive Graphs with R. Mol. Inform., 37.
- Love,M.I. et al. (2014) Differential analysis of count data - the DESeq2 package. Genome Biol., 15, 550.
- Robinson,M.D. et al. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140.
- Law,C.W. et al. (2014) Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol., 15.
- Young,M.D. et al. (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol., 11.
- Sergushichev,A. (2016) An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv, 60012.
